Welcome to the Research Group of Professor Mike Hannon
| Home | Research | Group Members | Publications | Collaborations | Joining the Group | Photos | Contact Us | Location | Links |
 |
Welcome to the Research Group of Professor Mike Hannon |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|||||||||||
| Press release: DNA Recognition | DNA Recognition | Cellular and Nuclear Targeting | Supramolecular Architectures |
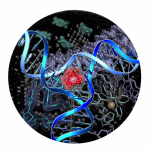
NMR structure of a cylinder in the major groove of DNA
Within biological systems, sequence specific DNA recognition is achieved by the surface motifs of proteins, which generally interact non-covalently with the major groove of DNA. Such protein recognition of DNA contrasts with that of synthetic small molecule recognition agents. Being smaller in scale than proteins, they tend to target the minor groove or act via intercalation.
The concept we have pioneered is that supramolecular chemistry should allow construction of large synthetic structures that closely mimic the dimensions of protein DNA recognition motifs, that such agents should have novel DNA-binding properties as a result, and hence should display new types of activity in biological systems. In particular our initial focus was to create agents that would be the right size and shape to bind in the DNA major groove. In this way we would move beyond established synthetic intercalators and minor groove binding drugs.
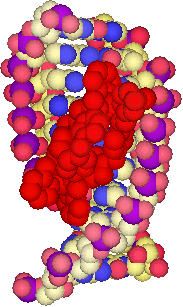
We designed a tetracationic triple-stranded cylinder (a helicate) ~2nm in length and ~1nm in diameter. These dimensions are similar to those of the alpha-helical DNA recognition unit of zinc fingers which bind in the major groove of DNA. Importantly, the size is too great to fit readily into the minor groove.
A tetracationic triple-stranded supramolecular cylinder
The agent not only binds to the major groove but in addition causes unexpected and dramatic intramolecular DNA coiling, which gives rise to small coils of DNA. Such coiling is unprecedented with synthetic agents and somewhat reminiscent of the effect of histones. By stepping up to the size-scale of nature, quite remarkable and new effects are observed.

AFM images showing the dramatic intra-molecular coiling induced by the cylinders
Proc. Natl. Acad. Sci., USA., 2002, 99, 5069-5074
Angew. Chem., Intl. Ed., 2001, 40, 880-884.
Still more remarkable is a second mode of binding discovered when the cylinder was crystallised with a palindromic hexanucleotide in a collaboration with Miquel Coll. The crystal structure revealed the cylinder to be located at the heart of a DNA 3-way junction (3WJ). This unexpected DNA structure is possible because, for a palindromic DNA, Watson-Crick hydrogen bonding can be satisfied either through a duplex structure, or through junction structures (e.g. 3WJ, 4WJ, 5WJ). The six phenyl rings of the cylinder face-face pi-stack onto the 6 DNA bases that are placed at the junction point, and the tetracation sits at the junction point where significant anionic charge is located. The cylinder and the junction are almost perfectly matched, like a hand and a glove, with neither significantly perturbed by the binding. NMR studies in a collaboration with Einar Sletten, established that this structure is not a crystallographic artefact but is the exclusive solution species formed when the cylinder and a palindromic DNA (the hexamer or a dodecamer) are mixed.
This DNA binding mode, a perfect shape-fit in the heart of a DNA junction, is unprecedented and quite different from those modes identified previously (intercalation, groove-binding, backbone-binding and alkylation) which dated back to the1960s.

A new mode of DNA binding; recognition of the centre of the 3-way DNA junction
Angew. Chem., Intl. Ed, 2006, 45, 1227-1231. Featured on the journal front cover
The first new type of DNA recognition by a drug for forty years! For more details of this work and its importance follow this link...
Y-shaped DNA junction structures are particularly exciting as they are found whenever DNA replicates itself (the replication fork). An ability to bind (and stall) a replication fork could be used to stop cancer cells dividing. We have shown that these cylinders have similar levels of cytotoxic activity against cancer cell lines as the clinical platinum drugs (which form metal-ligand coordination bonds to the bases). However their molecular level action on the DNA is completely different from that of cisplatin. Whereas cisplatin is genotoxic and a mutagen (clinically this is very undesirable!) the cylinders with their non-covalent recognition of DNA are non-genotoxic and non-mutagenic. They are powerful cytostatic agents arresting cells in the G0/G1 phase of the cell cycle and subsequently inducing death through apoptosis.
Chemistry & Biology, 2008, 15, 1258-1267
Angew. Chem., Intl. Ed., 2007, 46, 4374-4378
We have also explored whether we could push the approach further and combine the cylinder design with a cisplatin-type design to afford still better activity. In the cylinders used above the metal centers are fully coordinated by the bridging ligands; these are termed "saturated" cylinders (or "saturated" helicates). To combine the two types of design, we developed "unsaturated" double-stranded analogues, in which there are vacant coordination sites that offer the additional possibility of the metal center interacting directly with DNA. We selected ruthenium, rather than platinum, as our metal center of choice for its higher coordination number. An azo analogue of the imine ligand used to create the saturated cylinders enabled different isomers of the unsaturated [Ru2Cl4L2] compounds to be isolated. The compounds are very active against cancer cell lines: with their potencies ranging from being similar to that of cisplatin to being as much as 100 times more potent. Further biological studies are in progress to fully understand and correlate the biological activity and DNA binding of these new classes of complex.

An unsaturated [Ru2Cl4L2] supramolecular helicate which is active against cancer cell-lines
Angew. Chem., Intl. Ed., 2006, 45, 4839-4842
Current research on these supramolecular helicates centres around functionalising the cylinder for biological applications while mainting the DNA recognition properties. This is allowing us to investigate its effects on cellular morphology and function in cancer cell lines.
 |
 |
G-quartets
(left) stack up giving a G-quadruplex (right)
This project benefits from a range of European collaborations and the work has received extensive international media coverage and been highlighted in Scientific American.
For a review on non-covalent DNA recognition see:
Chem. Soc. Rev., 2007, 36, 280-295